DNA nanotechnology
Artificial cytoskeleton made of DNA for synthetic cells
The physicists Prof. Dr. Kerstin Göpfrich and Prof. Dr. Laura Na Liu want to understand life from the bottom up. They intend to do this by constructing an artificial cell. However, rather than natural protein building blocks, they are using three-dimensional DNA structures as construction material. The first step involved successfully creating an artificial cell skeleton that dynamically assembles and disassembles like the biological model and can transport vesicles.
 The two physicists Prof. Dr. Laura Na Liu (left) from the University of Stuttgart and Prof. Dr. Kerstin Göpfrich (right) from the ZMBH are cooperating closely to develop the basics of a synthetic model cell. © University of Stuttgart (left) I Katrin Binner on behalf of MPG (right)
The two physicists Prof. Dr. Laura Na Liu (left) from the University of Stuttgart and Prof. Dr. Kerstin Göpfrich (right) from the ZMBH are cooperating closely to develop the basics of a synthetic model cell. © University of Stuttgart (left) I Katrin Binner on behalf of MPG (right)"As a physicist, I like to think in models. My research group is developing a synthetic model cell that reconstructs the functions of a biological cell in a simplified way," says Prof. Dr. Kerstin Göpfrich from the Centre for Molecular Biology at Heidelberg University (ZMBH), explaining her scientific approach. The researcher, who also heads up a group at the Max Planck Institute (MPI) for Medical Research, wants to use this strategy to construct a living cell from scratch in the long term. Together with Prof. Dr. Laura Na Liu’s research group from the 2nd Physics Institute at the University of Stuttgart, Kerstin Göpfrich’s team has already successfully undertaken a first step to produce functional, artificial filaments that fulfil elementary properties of the cell skeleton. "We use DNA as a material to create precise three-dimensional structures and thus understand fundamental physical processes," explains Liu, a fellow at the MPI for Solid State Research.
Smallest components using DNA nanotechnology
In living cells, deoxyribonucleic acid (DNA) carries genetic information and is neatly packaged in the cell nucleus in the form of a double-stranded helix. The backbone of the DNA strands is made of alternating molecules of the sugar deoxyribose and phosphate groups. One of four bases (adenine, cytosine, guanine or thymine) is attached to each sugar molecule. The order, i.e. the sequence of the bases along the DNA backbone encodes the genetic information.
In a DNA double helix, each base interacts with its matching, complementary partner on the opposite strand. Adenine is paired with thymine, and cytosine is paired with guanine. In single-stranded DNA, domains can fuse together when they contain complementary sequences. The driving force behind this process is the molecule’s energy state. The greater the number of complimentary base pairs formed, the more energetically favourable this is.
Back in the early 1980s, the biochemist and crystallographer Nadrian Seeman realised that single-stranded DNA pieces fold independently and predictably depending on their sequence. He also found that different strands can stably pair with each other. Three-dimensional structures form if the sequences are combined intelligently. In 1991, Seeman constructed the first three-dimensional nanoscale object, a cube made of DNA, thus laying the foundations for DNA nanotechnology.
At the beginning of the new millennium, the computer scientist Paul Rothemund developed a technique to manipulate and fold strands of DNA known as DNA origami, which built on Seeman’s work. Rothemund’s technique involves the reliable folding of long DNA strands – mostly of viral origin – into arbitrary complex structures with the help of short synthetic auxiliary strands.
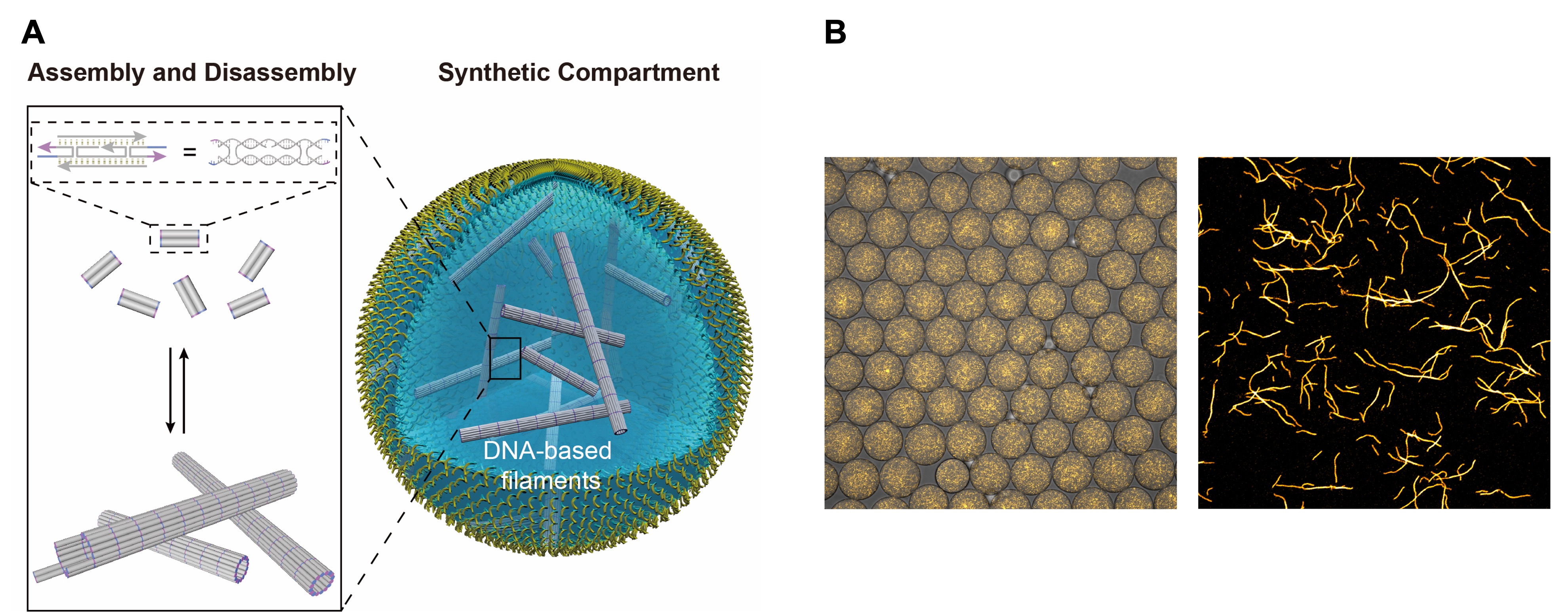 A Five different pieces of DNA assemble into a "tile", several thousand of which form a tubular filament. The assembly and disassembly can be triggered by external stimuli. B Microscope image of the DNA-based filaments labelled with a fluorescent dye.
Source: A https://www.nature.com/articles/s41557-022-00945-w, Laura Na Liu and Kerstin Göpfrich, CC-BY 4.0 (http://creativecommons.org/licenses/by/4.0/), B https://www.nature.com/articles/s41557-022-00945-w, Laura Na Liu and Kerstin Göpfrich, CC-BY 4.0 (http://creativecommons.org/licenses/by/4.0/), modified by R. Menßen-Franz
A Five different pieces of DNA assemble into a "tile", several thousand of which form a tubular filament. The assembly and disassembly can be triggered by external stimuli. B Microscope image of the DNA-based filaments labelled with a fluorescent dye.
Source: A https://www.nature.com/articles/s41557-022-00945-w, Laura Na Liu and Kerstin Göpfrich, CC-BY 4.0 (http://creativecommons.org/licenses/by/4.0/), B https://www.nature.com/articles/s41557-022-00945-w, Laura Na Liu and Kerstin Göpfrich, CC-BY 4.0 (http://creativecommons.org/licenses/by/4.0/), modified by R. Menßen-Franz
What characterises a living cell?
Liu is an expert in the field of DNA origami technology and has already constructed different nanorobots that can perform turning, sliding or walking movements triggered by an external stimulus. She explains: "We have imitated the most diverse movements of cell molecules. We are now trying to integrate these artificial components into synthetic cells. If we really want to understand how a cell works, we have to build it from scratch."
When she was working on her doctoral thesis, Göpfrich specialised in the incorporation of artificial DNA elements into vesicles with the ultimate goal of creating a living cell. She comments: "Natural components that have evolved over millions of years are very complex. It is far easier to model life using an abstract way of thinking that produces new elements." She believes that a synthetic cell may well be designed differently from its natural model. Genetic information, i.e. DNA, does not necessarily have to be transcribed into RNA and then be translated into proteins. Other solutions are also conceivable if one considers the ability to self-replicate and evolve to be the minimum requirement of living systems. "DNA nanotechnology makes it possible to design very precise components, often with more scope than protein engineering can currently offer. Moreover, we have a direct link between information and function, so we don't need transcription and translation."
Artificial cytoskeleton made of DNA filaments
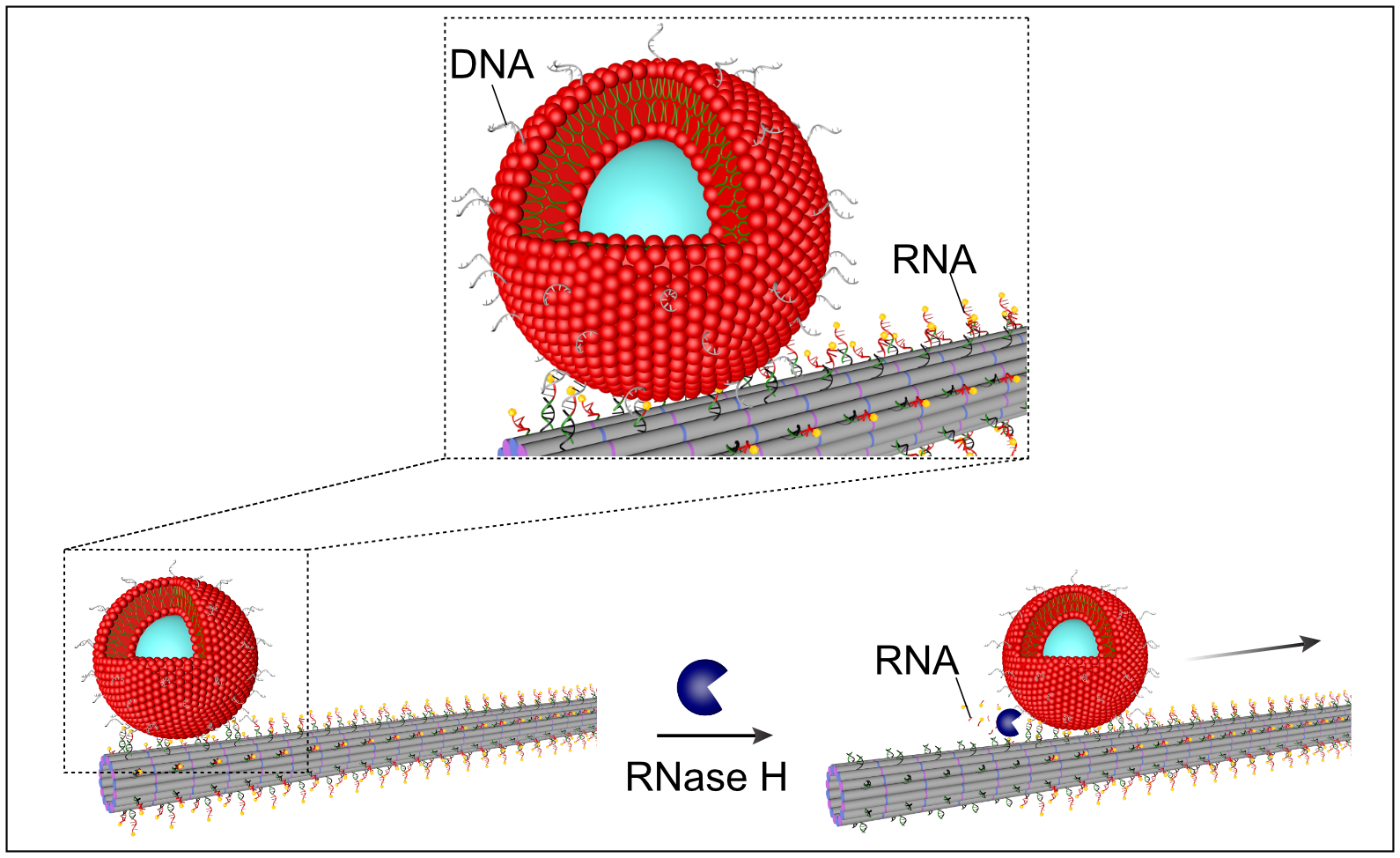 DNA pieces on the surface of the vesicles hybridise with RNA pieces on the filaments. The enzyme RNase H cuts both the bond and the RNA involved, initiating a rolling movement of the vesicles along the filaments. Source: https://www.nature.com/articles/s41557-022-00945-w, Laura Na Liu and Kerstin Göpfrich, CC-BY 4.0 (http://creativecommons.org/licenses/by/4.0/), modified by R. Menßen-Franz.
DNA pieces on the surface of the vesicles hybridise with RNA pieces on the filaments. The enzyme RNase H cuts both the bond and the RNA involved, initiating a rolling movement of the vesicles along the filaments. Source: https://www.nature.com/articles/s41557-022-00945-w, Laura Na Liu and Kerstin Göpfrich, CC-BY 4.0 (http://creativecommons.org/licenses/by/4.0/), modified by R. Menßen-Franz.The two skilled physicists met in Stuttgart, where Göpfrich had worked at the MPI for Intelligent Systems for several years. Together with doctoral student Kevin Jahnke and postdoctoral researcher Dr. Pengfei Zhan, they successfully reconstructed the basic structure of a eukaryotic cell – the cytoskeleton. For this purpose, they used known DNA pieces that were only 50 base pairs long and encapsulated them in cell-like compartments using microfluidic methods. Five different DNA strands form a so-called "tile". Hundreds of these "tiles" can cluster together and form tubular filaments. "We have the DNA fragments produced synthetically, mix them together in the laboratory and heat them. When they cool down, these fragments then strive for the most energetically favourable state and form filaments," Göpfrich says, describing the process. Liu adds: "The filaments are several micrometres long, but only ten to twelve nanometres in diameter." This makes them similar to microtubules in cells, which are also made up of many small elements known as tubulin molecules.
The natural cytoskeleton gives the cell its shape, but is also responsible for transporting molecules within the cell and for movement. It constantly assembles and disassembles, depending on the surrounding stimuli. After adding further elements, the researchers were also able to reproduce this property, as well as directing growth from a starting segment. Cytoskeleton assembly was triggered by the physiological energy carrier ATP (adenosine triphosphate), just like in cells, and the growth dynamics of the artificial system corresponded to the natural filaments, even if growth wasn’t yet taking place under ATP consumption.
According to Göpfrich, reproducing the complex transport function is the most challenging part of the group’s research, which was published in the renowned journal Nature Chemistry in August 2022.1) For this purpose, the team attached short pieces of RNA to the surface of the filaments. The sequences of these RNA pieces were complementary to pieces of DNA on the "cargo", in this case small vesicles. RNA-DNA hybrids form, thus enabling the vesicles to bind to the filaments. The addition of the enzyme RNase H, which breaks down this bond as well as the RNA involved, causes DNA pieces to have to find new RNA partners elsewhere in the vesicle, resulting in a directed rolling motion along the filaments. "This work is very promising, even though we cannot yet determine the starting direction and the process is relatively slow," Liu says. Göpfrich adds: "Interestingly, the influenza virus rolls itself across a cell surface in a very similar way. So we have now created a model system that we can use to study such transport mechanisms in the future."
Great potential of blue sky science
The results illustrate that research where clear applications are not immediately apparent and which is driven primarily by researchers’ curiosity (so-called blue sky science) can produce unexpected but very useful things. "The methods we are developing as we work towards producing an artificial cell are often very application-oriented and can be used directly with medical doctors and biologists to address relevant issues," Göpfrich says enthusiastically. "Our vision is to create life as we don't know it. But in so doing, we can also learn a lot - often in unexpected ways - about existing life forms." "Our approach also opens up the possibility of going beyond the functions of natural biological systems," Liu points out.