Computer-assisted genome mining
Natural product genomics opens up new avenues in the search for antibiotics
Antibiotic-resistant pathogens are increasingly endangering our health. Since most of the drugs currently in use are based on secondary metabolites produced by bacteria or fungi, the research group of Prof. Dr. Nadine Ziemert in Tübingen is developing bioinformatic tools to specifically search the genome of these organisms for previously unknown antimicrobial agents.
Many bacteria, fungi and plants produce not only the substances necessary for growth, development and reproduction, but also bioactive compounds as part of the so-called secondary metabolism. These non-essential, low-molecular secondary metabolites have a wide variety of tasks. Some act as signalling or communication substances, or as protection against being eaten by other organisms. Others specifically inhibit the reproduction of competing and pathogenic microorganisms in their immediate environment or even kill them.
Most antibiotics used in medicine today, such as penicillins, streptomycin, chloramphenicol and tetracyclines, are based on this type of natural substance, which is produced predominantly by bacteria and fungi. These substances are highly complex compounds that consist of unusual building blocks and can only be synthesised with the help of specialised enzymes. The molecules block fundamental processes in the target organism and are often effective against several species of bacteria (broad-spectrum antibiotics). Since they attack structures or mechanisms that do not exist in human cells, they are usually well tolerated by humans.
New antibiotics are urgently needed
 Prof. Dr. Nadine Ziemert from the University of Tübingen is using natural product genomics to search for new antibiotics. © University of Tübingen
Prof. Dr. Nadine Ziemert from the University of Tübingen is using natural product genomics to search for new antibiotics. © University of TübingenHowever, due to the widespread use of antibiotics over many decades, pathogens are now increasingly developing resistance to them. Infections with multidrug-resistant bacteria can lead to severe physical damage and, according to the latest estimates from the World Health Organisation (WHO), cause 1.3 million deaths worldwide every year.1)
"It is already five to twelve; we urgently need new antibiotics. And since the traditional approach is very lengthy, we need to take other paths," says Prof. Dr. Nadine Ziemert from the University of Tübingen. Ziemert conducts research at the interface of microbial ecology, microbiology and bioinformatics, and her research group at the Interfaculty Institute for Microbiology and Infection Medicine develops computer-based methods to make the identification of new antimicrobial natural products faster and more effective. She comments: "We are far from having exhausted the potential of bacterial secondary metabolites. Over the course of evolution, nature has optimised the mechanisms of action and we should take advantage of this."
Identifying relevant biosynthetic gene clusters through natural product genomics
Most antibiotics in use today originate from soil-dwelling actinobacteria, particularly streptomycetes. The standard way to search for new substances is therefore to grow organisms from soil samples in the laboratory and test the effect of the substances they produce on known pathogens. Unfortunately, over the course of the investigations it often turns out that the compounds are already known. As this is a very costly process and the financial returns are low, the pharmaceutical industry withdrew from antibiotic research years ago.
Ziemert and her team have chosen the novel approach of genome mining (natural product genomics) to analyse the information available in the bacterial genome. Since the Haemophilus influenzae genome was the first microorganism to be sequenced in 1995, the amount of data has increased rapidly. The biologist and bioinformatician explains: "If I know how to read the DNA, I can also identify substances that are only produced under specific conditions. On the computer, I can therefore examine organisms that have never been cultivated in the laboratory. The big challenge is to develop algorithms and databases that can filter out the relevant information from huge amounts of data."
The advantage for the researchers is that so-called biosynthetic gene clusters (BGCs) are almost always responsible for the synthesis of a secondary metabolite. BGCs are collections of sometimes more than 75 genes that are all located in close proximity in the genome. Antimicrobial natural compounds often belong to the class of non-ribosomal peptides (amino acid chains that are not formed at the ribosome in the cell as is usually the case) or to the polyketides (formed from acetate units). They are synthesised by large, modular multi-enzyme complexes known as nonribosomal peptide synthetases and polyketide synthases. Since the underlying genes contain very specific sequences, the relevant BGCs can now be quickly identified using bioinformatic methods. "However, it is then difficult to distinguish whether bacteria with similar gene clusters produce the same natural product or compounds of different classes," explains Ziermert, who is also a Professor of Translational Natural Product Genomics.
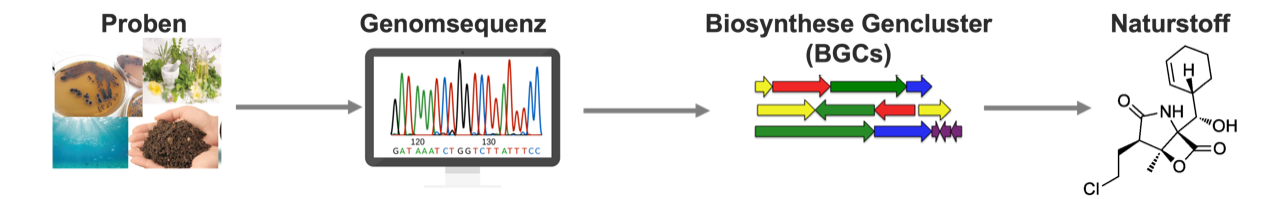 The genome sequence of bacteria and fungi from a wide variety of samples is used to identify the biosynthetic gene clusters responsible for producing a natural substance. © University of Tübingen
The genome sequence of bacteria and fungi from a wide variety of samples is used to identify the biosynthetic gene clusters responsible for producing a natural substance. © University of Tübingen
Many unexplored secondary metabolites present in bacteria
PhD student Athina Gavriilidou analysed almost 170,000 genomes of cultivated and previously sequenced bacteria to determine the breadth of chemical diversity in the natural compounds produced by bacteria and to identify the most promising organisms in the search for new antibiotics. She identified more than 50,000 different classes of natural compounds, of which only about 2,500 are known. The data published last year in the renowned journal Nature Microbiology2) illustrate the extent of the hidden potential and also define previously unknown "hotspots" among bacterial genera, which are genetically very diverse and therefore promising in the search for new active substances.
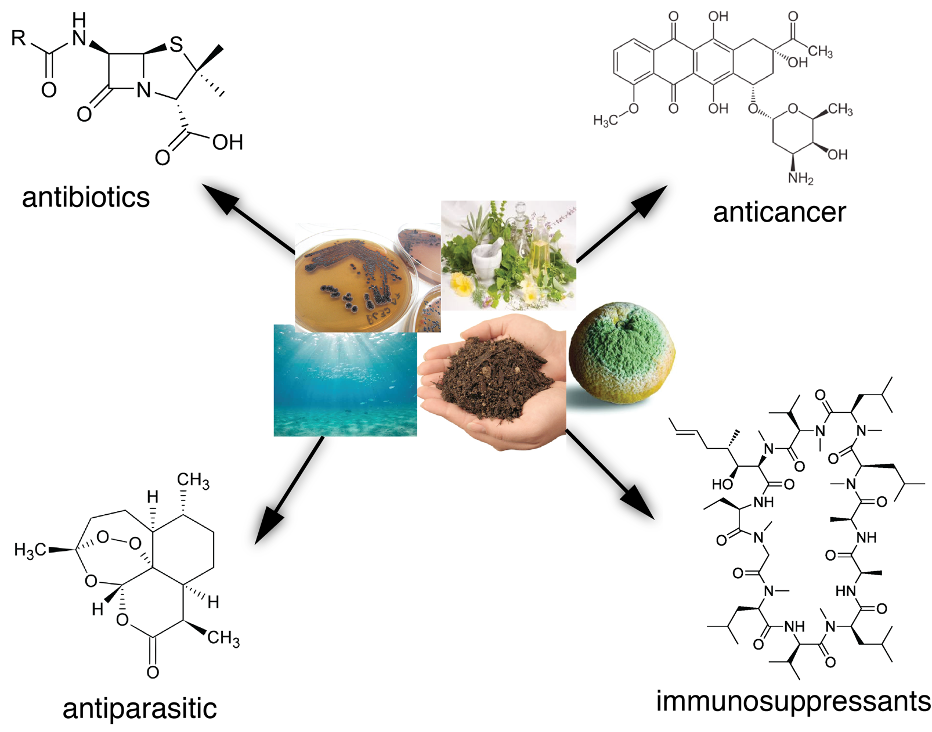 Some of the secondary metabolites produced in bacteria and fungi of different origins can act as antibiotics, anticancer agents, immunosuppressants or may be effective against parasites. © University of Tübingen
Some of the secondary metabolites produced in bacteria and fungi of different origins can act as antibiotics, anticancer agents, immunosuppressants or may be effective against parasites. © University of TübingenIn addition, the researchers looked at the genetic material from various environmental samples. This so-called metagenome is obtained directly from samples without requiring cultivation in the laboratory and contains the genomes of all bacteria present in a sample. It turned out that half of the classes of natural compounds present were completely unknown, demonstrating that it is worth searching for effective antimicrobial agents directly in the environment.
The interdisciplinary working group, which is part of the German Centre for Infection Research (DZIF) and also affiliated with the Interfaculty Institute for Biomedical Informatics, has already identified some interesting BGCs. The next challenge is to cultivate the corresponding bacteria and isolate and characterise the active substances. The researchers are working closely with their colleagues from the Tübingen Cluster of Excellence CMFI (Controlling Microbes to Fight Infections). "Our method also takes time, but we are working under better conditions than simple random searches," Ziemert says.
Broad application of natural product genomics
Basically, Ziemert is interested in all secondary metabolites and wants to understand which substances are produced under which conditions in certain biotic communities and how they interact. Natural products are increasingly proving to be important components of complex ecological systems. "With this knowledge, we could expand the concept of antibiotics in the future and possibly use secondary metabolites to strengthen the local microbiome in advance so that pathogenic organisms have less of a chance. Whether it’s about making plants resistant to germs or supporting the immune-modulatory effects of our gut microbiome," says Ziemert, a pioneer of natural product genomics with a clear vision.
Since many secondary metabolites are also harmful to their producers, BGCs often contain genes whose protein products mediate protection. These can be very unspecific transporters that move the active substance out of the cell, or enzymes that destroy it. Sometimes, however, they are also modified versions of the target protein to which the substances cannot bind. In such cases, the gene clusters contain additional important information about the mechanism of action. Targeted genome mining takes advantage of this and uses the sequence of a pre-defined target (target protein) to specifically search for BGCs that are responsible for producing a relevant substance. This method can be applied to all natural products and can also be used in the search for new cancer drugs, analgesics or immunosuppressants.