Computer tool opens up new ways to address antibiotic resistance
PhARIS identifies effective phages against multi-resistant bacteria
Bacteriophages - viruses that specifically attack and destroy bacteria - are emerging as promising alternatives to antibiotics in the fight against drug-resistant infections. To speed this effort, researchers at the University of Tübingen have created PhARIS, a computational tool that mines genomic data to pinpoint phages that are effective against the hospital superbug Staphylococcus aureus. By rapidly matching phage and pathogen, PhARIS can significantly accelerate the development of targeted phage therapies.
Drugs are increasingly losing their effectiveness against bacteria, viruses and even fungi. This growing antimicrobial resistance (AMR) is considered one of the most pressing threats to global health today. According to estimates from the international research consortium Global Burden of Disease (GBD), bacterial AMR alone was linked to between 4 and 7.1 million deaths worldwide in 2021.¹) Particularly alarming is the rise of methicillin-resistant Staphylococcus aureus (MRSA), which now accounts for over 11 percent of fatal infections.¹)
Under normal conditions, gram-positive, spherical S. aureus bacteria are harmless. In about one-third of the population, they colonise the nose or skin as part of the natural microbiome. However, when the immune system is weakened or the skin barrier compromised, these bacteria can cause not only local infections but also serious, potentially life-threatening conditions such as sepsis, endocarditis or toxic shock syndrome. Alarmingly, around 80% of S. aureus strains are now resistant to penicillin.²) In cases where resistance extends to other β-lactam antibiotics - such as methicillin or clindamycin - and to drugs from other classes, including erythromycin or quinolones, the resulting multidrug-resistant strains become extremely difficult to treat. MRSA (methicillin-resistant Staphylococcus aureus) is most commonly found in healthcare settings and accounts for over 20% of all S. aureus infections in Germany.²)
With phages against bacteria
"The numbers keep rising, and we urgently need alternative treatment strategies," says Professor Dr. Andreas Peschel from the Interfaculty Institute of Microbiology and Infection Medicine at the University of Tübingen (IMIT). Together with other research groups within the ‘Control of Microorganisms to Fight Infections (CMFI)’ Cluster of Excellence he is working to develop innovative approaches for the precise elimination of specific pathogens. "Most current antibiotics have a broad spectrum of activity and inevitably harm our beneficial gut microbiome. Our goal is to design highly specific solutions that target only harmful bacteria," he explains.
 A team led by Professor Dr. Andreas Peschel (left) and doctoral researcher Janes Krusche (right) from the Interfaculty Institute of Microbiology and Infection Medicine (IMIT) at the University of Tübingen has developed a computer tool called PhARIS, which can be used to identify specific phages targeting the hospital pathogen S. aureus. © Jörn Jäger/University of Tübingen (left); Leon Kokkoliadis/University of Tübingen (right)
A team led by Professor Dr. Andreas Peschel (left) and doctoral researcher Janes Krusche (right) from the Interfaculty Institute of Microbiology and Infection Medicine (IMIT) at the University of Tübingen has developed a computer tool called PhARIS, which can be used to identify specific phages targeting the hospital pathogen S. aureus. © Jörn Jäger/University of Tübingen (left); Leon Kokkoliadis/University of Tübingen (right)Peschel and his team are working on bacteriophages, i.e. special viruses that can only infect and kill bacteria. The antibacterial properties of a novel substance were described as early as 1915 by the Englishman Frederik Twort and two years later by the French-Canadian Félix D'Hérelle.3) Despite several successful treatments with the viruses known as bacterivores (from the ancient Greek phagein = to eat), phage therapy became increasingly forgotten with the advent of antibiotics in Western countries. However, in the former Soviet states, Georgia in particular, the treatment method was further developed and is still widely and very successfully used today.
In light of rising antibiotic resistance, there is growing global interest in phage therapy. However, there are a number of challenges associated with their use. Biologist Professor Andreas Peschel explains: "Phages are significantly larger than antibiotics and cannot penetrate tissues as easily. This limits their application mainly to superficial infections or to infections accessible via the bloodstream. They are also propagated in bacteria, which makes it difficult to produce high quality phages with any degree of consistency. This is a major barrier to regulatory approval as medicinal products. Most notably, phages are highly specific and target only certain bacterial species or specific subtypes."
PhARIS analysis programme finds suitable phages
 PhARIS (Phage Aureus RBP Identification System) analyses the genetic information of phages to identify their receptor-binding proteins and thereby finds suitable candidates to infect and destroy S. aureus. © Dr. Johannes Richers and Dr. Christoph Kuehne (Jo Richers Studio), commissioned by the University of Tübingen/CMFI
PhARIS (Phage Aureus RBP Identification System) analyses the genetic information of phages to identify their receptor-binding proteins and thereby finds suitable candidates to infect and destroy S. aureus. © Dr. Johannes Richers and Dr. Christoph Kuehne (Jo Richers Studio), commissioned by the University of Tübingen/CMFIThe high specificity of these viruses protects the human microbiome but also makes their routine use extremely challenging. Until now, once a pathogen is isolated, identifying the appropriate phage has involved a time-consuming trial-and-error process that can take days or even weeks, due to the hundreds of potential candidates. PhD student Janes Krusche has now developed PhARIS (Phage Aureus RBP Identification System), an analytical programme that predicts which S. aureus variants a phage can infect based solely on the virus’s genetic information.
Almost all phages consist of a head segment containing genetic material - either DNA or RNA - and a tail segment that carries receptor-binding proteins (RBPs). These RBPs specifically recognise and bind to certain variants of the sugar-containing components on the bacterial cell wall. After irreversible attachment, the phage injects its genetic material into the bacterial interior and hijacks the host’s metabolic machinery in order to replicate. Within a short period (typically 20 to 30 minutes), the infected cell bursts, releasing new viral particles that can infect and destroy other bacteria.
Krusche and his colleagues identified and analysed the receptor-binding proteins (RBPs) and their associated genes from a total of 335 phages that infect S. aureus. They also elucidated the molecular mechanisms underlying the binding to different pathogen strains and used this knowledge to develop the PhARIS computer tool. Krusche, the first author of a paper published in the scientific journal CellReports4), emphasises: "It is now possible to predict which S. aureus clones a phage can target using only its genetic information. This significantly accelerates the development of phage therapies."
Phage therapy as part of the solution
The analysis programme is freely accessible worldwide and has huge potential to advance the broader use of phage therapies. In Germany, efforts to establish this treatment approach are also gaining momentum. For instance, the Phage4Cure project is exploring the effectiveness of inhalable phage cocktails against severe pneumonia caused by multi-resistant Pseudomonas aeruginosa bacteria.5) Until now, only so-called compassionate-use trials with phages have been permitted in Germany. These are treatments that deviate from standard medical practice and can only be administered to patients with serious conditions where approved therapies have failed.
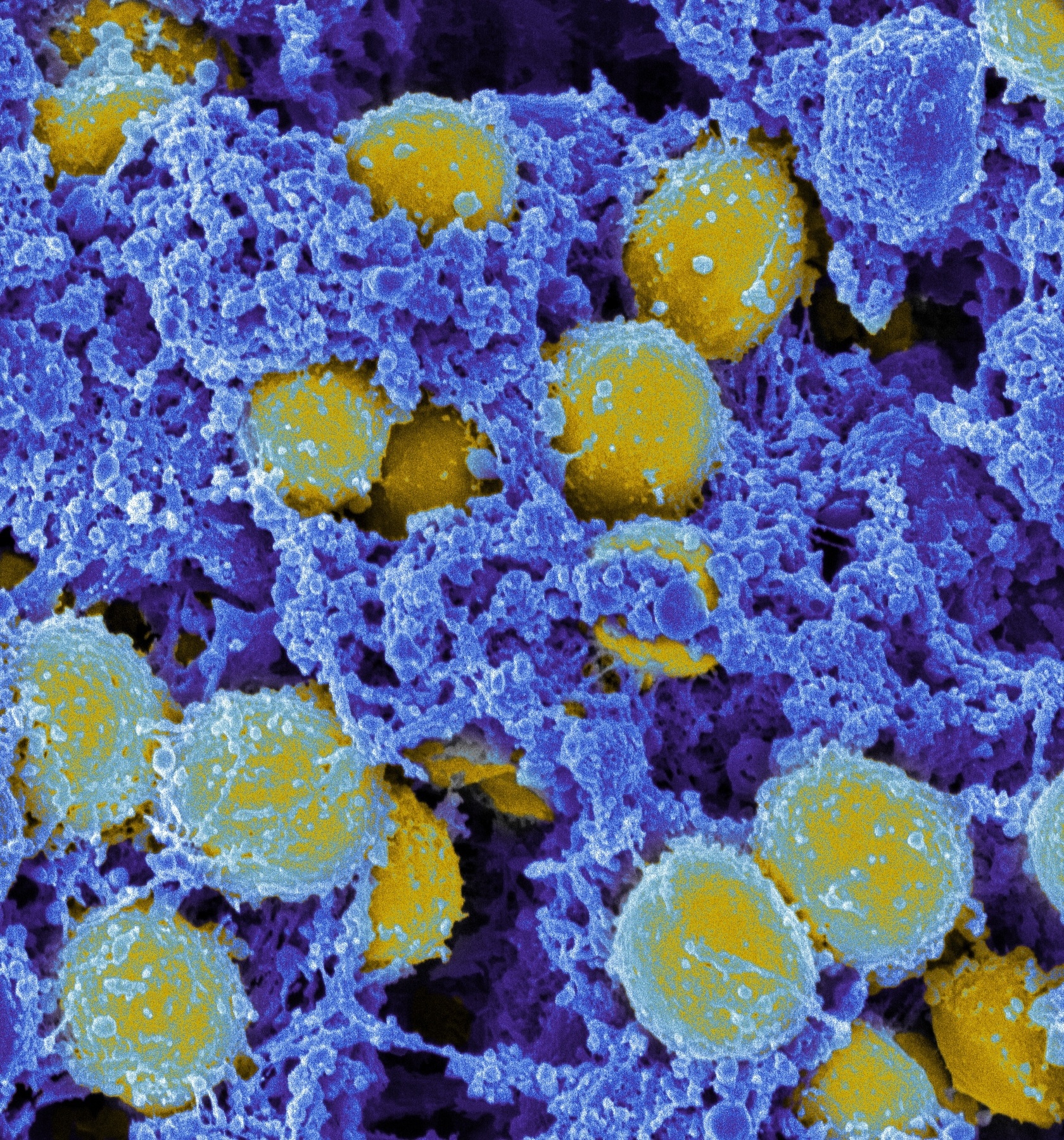 Scanning electron micrograph of S. aureus (yellow). Source: ‘Staphylococcus aureus Bacteria’ by NIAID, licensed through CC BY 2.0.(https://creativecommons.org/licenses/by/2.0/)
Scanning electron micrograph of S. aureus (yellow). Source: ‘Staphylococcus aureus Bacteria’ by NIAID, licensed through CC BY 2.0.(https://creativecommons.org/licenses/by/2.0/)PhARIS currently analyses only phages targeting S. aureus pathogens. However, with suitable genetic data, the system could potentially be adapted to other bacteria. The primary focus is on the group of multi-resistant ESKAPE pathogens, which are major causes of hospital-acquired infections.6)
However, as Peschel emphasises, "Phage therapies are not intended to replace antibiotics but for use as a complementary treatment. This is because phage therapy requires a complex, individualised approach, where both the pathogen subtype and the matching phage must be identified beforehand. Nonetheless, the strategy is highly promising, as the viruses disappear once the targeted pathogenic bacteria have been eliminated, since they can no longer replicate."
The researchers especially envision the use of phage therapy as a prophylactic treatment for high-risk patients, such as those undergoing planned surgical procedures. Since all ESKAPE pathogens naturally reside in the human microbiome - particularly in the nose and on the skin - targeted elimination of these bacteria before they become pathogenic due to a weakened immune system during medical treatment could significantly reduce the risk of serious infections.
With PhARIS, the researchers have now made an important contribution to progress in the fight against antibiotic-resistant pathogens. "But we are not medical practitioners, the actual application will be used by doctors."