Gene regulation as a starting point for cancer therapies
New investigation method for deciphering complex epigenetic networks
The development and maintenance of uncontrolled cell division in tumours is often due to the unbalanced, complex interplay of regulatory epigenetic networks. Researchers at the Institute of Biochemistry and Technical Biochemistry in Stuttgart have developed a new screening system to identify essential components that can serve as targets for anticancer drugs.
All our body cells contain identical genetic information, yet we possess more than 300 different cell types with different functions. The necessary differentiation during development from the fertilised egg cell to the organism is only possible through regulated gene transcription. This process is controlled by epigenetics, a superordinate level of regulation. It determines when and in which cells individual genes are used.
The 46 chromosomes in the nucleus of each cell are the carriers of genetic information. They consist of a long strand of nucleic acid that is condensed into chromatin with the help of special proteins called histones. "Epigenetics controls the degree of DNA packaging," explains Dr. Philipp Rathert, group leader at the Institute of Biochemistry and Technical Biochemistry (IBTB) at the University of Stuttgart. "Tightly packed DNA cannot be transcribed, whereas less tightly packed DNA regions are accessible to the transcription machinery." DNA transcription is influenced by small chemical modifications, such as the addition of acetyl residues (-C(O)CH₃) or methyl groups (-CH3) to the histones. These modifications primarily generate new binding sites for regulatory proteins and can both promote and inhibit transcription. Methylation of DNA building blocks (primarily cytosine) is another important mechanism that results in regulatory regions of genes (promoters, enhancers) being silenced.
Complex epigenetic networks regulate cell function
 Philipp Rathert, PhD, explores the complexities of the regulation of the epigenetic enzyme LSD1. © P. Rathert
Philipp Rathert, PhD, explores the complexities of the regulation of the epigenetic enzyme LSD1. © P. RathertDuring the development of an organism, epigenetic ‘tags’ are added to or removed from the DNA or histones, depending on the stage the cells are at. In the differentiated state, cell function then depends on the balanced interplay of these modifications and is passed on to the daughter cells during cell division. "When this dynamic system of regulation becomes unbalanced, cells can again de-differentiate and begin to divide uncontrollably. This is the first step towards the development of cancer," says Rathert describing the problem. His research group in the Department of Biochemistry studies, among other things, the enzyme lysine-specific histone demethylase LSD1, which plays an important role in a variety of cancers such as acute myeloid leukemia (AML) as well as breast, bladder and lung cancer. The enzyme is crucial for embryonic development and also plays an important role in maintaining the epigenetic balance in differentiated cells.
LSD1 as a target in cancer therapies
LSD1 is a component of various protein complexes and removes methyl groups from lysine 4 (K4) of histone H3. Depending on the complex partners, the removal of methyl groups at lysine 4 can either activate or switch off genes, but the exact regulation of the enzyme is still largely unclear. Since elevated levels of LSD1 occur in many cancers, and the laboratory finding that LSD1 inhibitors can reduce cancer growth, the latter are currently being investigated in clinical trials. The inhibitors in particular show good efficacy in AML and small-cell lung cancer (SCLC). "However, monotherapies often generate resistance in the long term and often have severe side effects due to the rather high dose required. That's why people are increasingly turning to combination therapies, in which several complex partners can serve as targets. This is where we started looking for essential interaction partners of LSD1," says the epigeneticist, explaining his laboratory's strategy.
Novel assay system to identify epigenetic coregulators
In order to shed light on the complex regulatory network surrounding LSD1, the scientist and his team first developed a fluorescence-based reporter gene system that can be used to analyse the activity of the LSD1 enzyme in living cells. Here, the genetic information for a red fluorescent protein (mCherry) is introduced into mouse cell lines under the control of a synthetic promoter with specific Tet repressor (tetR) binding sequences so that the cells glow intensely red. Furthermore, the cells produce the human LSD1 protein that is linked to the reverse tetR binding protein. Addition of doxycycline initiates the binding of LSD1, and thus its complex partners, to the promoter of mCherry. This alters the methylation of nearby histones and suppresses production of the fluorescent protein.
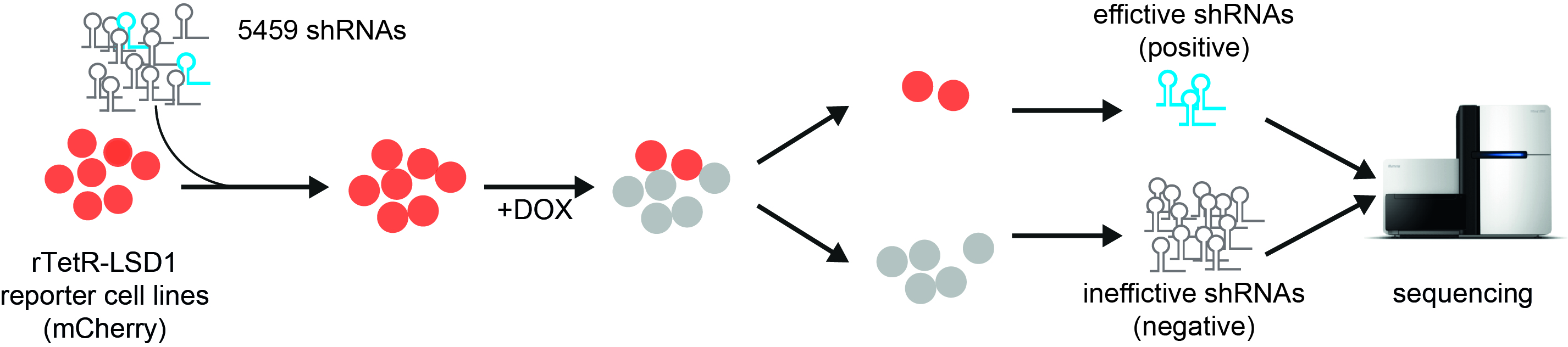 ChECS (chromatin effector coregulator screen) method: red fluorescent reporter cell lines producing LSD1 fused with the reverse tetR binding protein are transduced with a collection of shRNA molecules. Upon addition of doxycycline (DOX), LSD1 binds to the promoter of the mCherry gene and turns it off (grey cell lines). When a protein that is crucial for LSD1 function is switched off by shRNAs, the cells continue to glow red. The corresponding genetic information can be identified by sequence analysis. Source: “Workflow describing the ChECS screening“, P. Rathert, https://doi.org/10.1093/nar/gkab180, Oxford University Press, CC-BY-NC 4.0, modified by R. Menßen-Franz.
ChECS (chromatin effector coregulator screen) method: red fluorescent reporter cell lines producing LSD1 fused with the reverse tetR binding protein are transduced with a collection of shRNA molecules. Upon addition of doxycycline (DOX), LSD1 binds to the promoter of the mCherry gene and turns it off (grey cell lines). When a protein that is crucial for LSD1 function is switched off by shRNAs, the cells continue to glow red. The corresponding genetic information can be identified by sequence analysis. Source: “Workflow describing the ChECS screening“, P. Rathert, https://doi.org/10.1093/nar/gkab180, Oxford University Press, CC-BY-NC 4.0, modified by R. Menßen-Franz.The researchers then combined the reporter gene system with a functional genetic screen. This allows not only direct interaction partners to be identified, but also upstream proteins that can have an influence on enzyme activity too. In this case, the cells were transduced with a large number of different, short shRNA molecules. These form small hairpin structures and interact with the cell's mRNA molecules formed by gene transcription, preventing their translation into proteins. If the switched-off proteins are crucial for the function of LSD1, the cells continue to glow red even after doxycycline has been administered.
New regulatory cascade identified
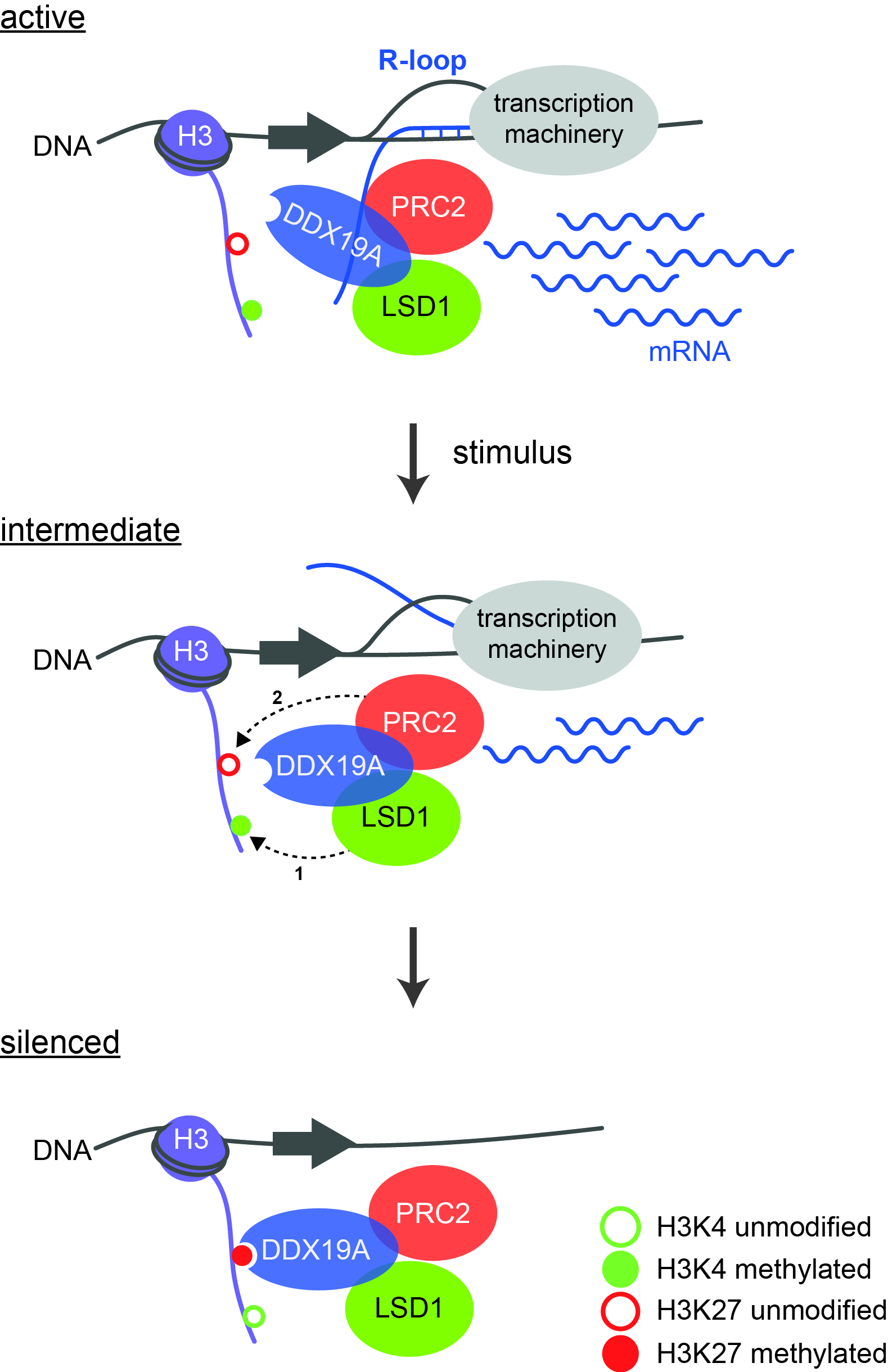 Model of the regulatory cascade leading to gene silencing. During active gene transcription, H3K4 is methylated and R-loops are formed. After an external stimulus has occurred, LSD1 demethylates H3K4 (1), whereupon PRC2 can bind and modify K27 (2). This creates a binding site for DDX19A, which completely removes the R-loops. Source: “Model of the regulatory cascade for transcriptional repression“, P. Rathert, https://doi.org/10.1093/nar/gkab180, Oxford University Press, CC-BY-NC 4.0, modified by R. Menßen-Franz.
Model of the regulatory cascade leading to gene silencing. During active gene transcription, H3K4 is methylated and R-loops are formed. After an external stimulus has occurred, LSD1 demethylates H3K4 (1), whereupon PRC2 can bind and modify K27 (2). This creates a binding site for DDX19A, which completely removes the R-loops. Source: “Model of the regulatory cascade for transcriptional repression“, P. Rathert, https://doi.org/10.1093/nar/gkab180, Oxford University Press, CC-BY-NC 4.0, modified by R. Menßen-Franz.With their innovative ChECS (chromatin effector coregulator screen) method, the group of researchers succeeded in identifying new LSD1 regulators in addition to already known ones. These include DDX19A, a helicase that can unwind special nucleic acid structures known as R-loops. These are RNA-DNA hybrids that form during transcription and also have regulatory effects. "We were able to show for the first time the connection between epigenetic modifications and the role of R-loops in gene inactivation," the biochemist reports. Active transcription is accompanied by the methylation of lysine 4 of histone H3 (H3K4me) and the formation of R-loops. Several steps are then required to completely silence the genes: after stimulation by an external signal, LSD1 removes the methyl groups from lysine 4 of histone H3. The polycomb repressive complex 2 (PRC2) can now bind and methylate K27, which on the one hand results in reduced transcription and on the other creates a binding site for the helicase DDX19A. The latter removes the remaining R-loops, which in turn stimulates the activity of LSD1 and PRC2. Robust silencing requires the activity of all components of the cascade.
Despite different initial mutations, many tumours share a similar transcriptional status that triggers uncontrolled cell division and is maintained by reprogrammed epigenetic machinery. For this reason, regulatory epigenetic networks are increasingly becoming the focus of cancer therapy. The novel investigation system developed by Rathert makes a decisive contribution to deciphering the complex relationships and can thus provide a basis for developing new combination therapies.